Unlocking the Secrets of Genetic Variation: A Deep Dive into SDR-seq
Unraveling the Impact of Coding and Noncoding Variants on Gene Expression
Summary
Scientists at the European Molecular Biology Laboratory (EMBL) have developed SDR-seq, a groundbreaking technology that can simultaneously decode both DNA and RNA from a single cell. This innovation finally opens a window into the noncoding regions of the genome the vast areas that harbor most disease-associated genetic variants. By revealing how these hidden changes influence gene activity, SDR-seq allows researchers to gain deeper insights into complex diseases and to develop more precise diagnostic tools.
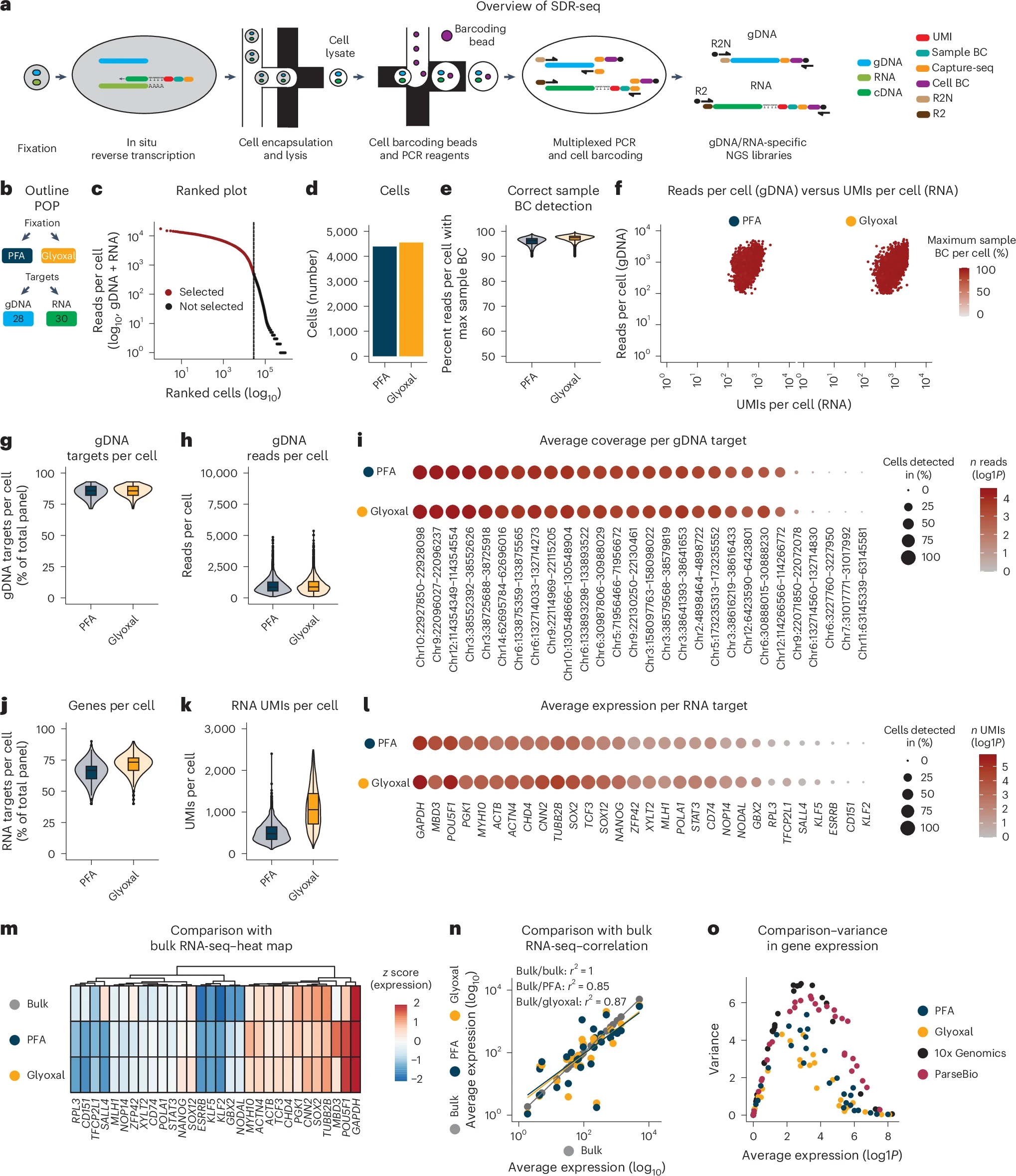
Fig. 1: SDR-seq links gDNA variants and gene expression in single cells.
a, Overview of targeted SDR-seq. R2N (Nextera) or R2 (TruSeq) overhangs on reverse primers enable separate NGS library generation for gDNA and RNA. b, Outline of the POP experiment. The fixation conditions and number of gDNA/RNA targets are indicated. c, Knee plot of ranked cells by sequencing depth (gDNA + RNA). d, Number of cells found per fixation condition. e, Correct sample BC detection per cell. Read count for maximum sample BCs found was divided by the total amount of RNA reads per cell; n = 4,391 (PFA) and 4,553 (glyoxal) cells from one SDR-seq experiment. f, gDNA reads per cell versus RNA UMIs per cell per fixation condition. Color indicates the percentage of reads per cell with max sample BCs. g,h, Number of gDNA targets per cell (g) and gDNA reads per cell (h); n = 4,391 (PFA) and 4,553 (glyoxal) cells from one SDR-seq experiment. i, Individual gDNA targets are shown per fixation condition. Size indicates the percentage of cells detected in. Color indicates read coverage; chr, chromosome. j,k, Number of genes per cell (j) and RNA UMIs per cell (k); n = 4,391 (PFA) and 4,553 (glyoxal) cells from one SDR-seq experiment. l, Individual genes are shown per fixation condition. Size indicates the percentage of cells detected in. Color indicates UMI coverage. m, Comparison of expressed genes to bulk RNA-seq data; z score data are scaled by row. n, Pearson correlation of expressed genes to bulk RNA-seq data. o, Average expression and variance of genes assayed in the POP experiment using SDR-seq, 10x Genomics and ParseBio.
Full Story
In the ever-evolving field of genomics, researchers are constantly seeking new tools to deepen our understanding of the complex interplay between genetic variants and gene expression. The recently developed single-cell DNA-RNA sequencing (SDR-seq) technique, described in the Nature Methods article "Functional phenotyping of genomic variants using joint multiomic single-cell DNA–RNA sequencing," offers a powerful solution to this challenge.
SDR-seq is a scalable and sensitive method that enables the simultaneous profiling of both genomic DNA (gDNA) and RNA targets in thousands of single cells. By linking precise genotypes to gene expression in their endogenous context, SDR-seq provides researchers with an unprecedented ability to study the impact of both coding and noncoding genetic variants.
Key Highlights of SDR-seq:
Targeted and Sensitive Approach: SDR-seq utilizes a targeted primer panel to achieve high coverage of gDNA and RNA targets, allowing for confident detection of genomic variants and their zygosity, as well as sensitive gene expression readouts. This contrasts with existing split-pooling or droplet-based approaches, which often result in sparse data and difficulties in correctly determining variant zygosity.
Scalability and Reproducibility:
The authors demonstrate that SDR-seq is scalable to detect hundreds of gDNA and RNA targets simultaneously, with high reproducibility and sensitivity across different panel sizes. This versatility enables the systematic study of both coding and noncoding variants, a crucial aspect given that the vast majority of disease-associated variants are located in the noncoding genome.
Linking Genotypes to Gene Expression:
SDR-seq's ability to directly measure gene expression changes associated with genetic variants sets it apart from other high-throughput approaches. The authors showcase this capability by probing the impact of strong and subtle gene expression changes across different perturbation systems, including CRISPRi, prime editing, and base editing.
Insights into B Cell Lymphoma:
By applying SDR-seq to primary B cell lymphoma samples, the researchers were able to link specific genetic variants to distinct gene expression patterns and cellular states, revealing the impact of mutational burden on tumorigenic signaling pathways.
The development of SDR-seq represents a significant advancement in the field of single-cell genomics, providing researchers with a versatile tool to unravel the complex regulatory mechanisms encoded by genetic variants. As the authors note, this method holds great potential to drive the development of therapeutic strategies and enhance our understanding of complex genetic disorders.
Conclusion:
The SDR-seq technique showcased in this Nature Methods article is a game-changer in the field of single-cell genomics. By seamlessly integrating the analysis of genomic DNA and gene expression in individual cells, it offers unprecedented insights into the functional consequences of both coding and noncoding genetic variants. As the scientific community continues to explore the vast potential of this technology, we can expect to see groundbreaking discoveries that will deepen our understanding of gene regulation and its implications for human health and disease.
Recent Posts
-
The Role of Technetium-99m in Sentinel Lymph Node Detection for Gynecological Cancers
The Role of Technetium-99m in Sentinel Lymph Node Detection for Gynecological Cancers Int …3rd Nov 2025 -
Unlocking the Secrets of Genetic Variation: A Deep Dive into SDR-seq
Unraveling the Impact of Coding and Noncoding Variants on Gene Expression Summary Scientists at the …21st Oct 2025 -
Current Bioinformatics Tools in Precision Oncology: An Integrated Review for Researchers
Current Bioinformatics Tools in Precision Oncology Abstract Precision oncology has transitioned from …14th Oct 2025